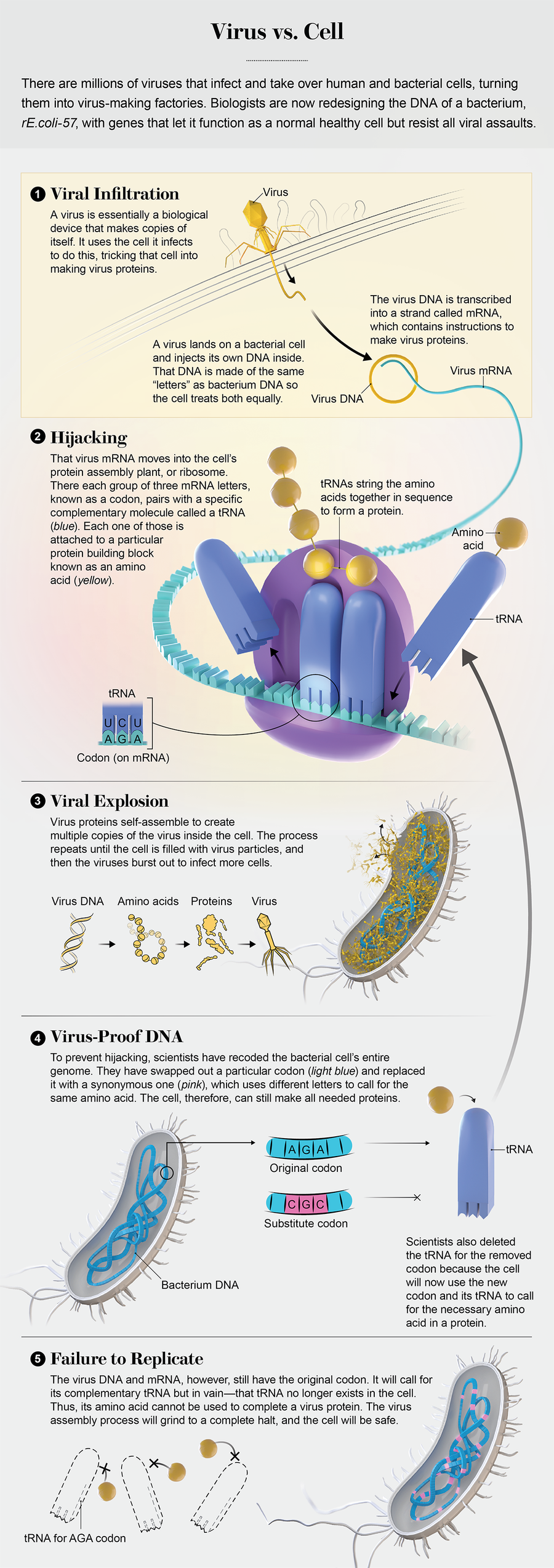
The virus touches down on the cell like a spider landing on a balloon 1,000 times its size. It has six thin legs splayed underneath a body that resembles a syringe with a bulbous head. This is a predator named lambda, and its prey is an Escherichia coli bacterium. Having found its victim, lambda now does what uncountable trillions of viruses have done since life first emerged: it latches onto the cell membrane with its legs, attaches its syringelike part to a pore and contracts, injecting its DNA inside.
The DNA contains the instructions for making more viruses, and that is pretty much all a virus is: a protein capsule holding blueprints for building more copies of itself. Viruses do not have the molecular machinery to build new things. Instead they break into cells and hijack cellular equipment, using it to replicate until there are so many viruses, they burst through the cell walls. They can do this because all organisms, from rhinoceroses on African plains to rhinoviruses infecting your nose, use the same coding system, which is based on nucleic acids such as DNA. Feed the code into the cell, and it will use those instructions to build proteins.
In the infected bacterium, that process starts. New viral proteins take shape. Things are looking good for lambda. Within minutes the cell will be bursting at the seams with a multitude of brand-new viruses. When they break out, each one will head for another bacterium, aiming to repeat this cycle over and over again.
On supporting science journalism
If you're enjoying this article, consider supporting our award-winning journalism by subscribing. By purchasing a subscription you are helping to ensure the future of impactful stories about the discoveries and ideas shaping our world today.
Then the cellular machinery freezes. It simply cannot read the virus’s DNA. In the seemingly eternal duel between virus and cell, this failure has never happened. And now it means lambda is doomed.
The reason for its demise is that this strain of E. coli has been reprogrammed to use a DNA operating system that has never existed on earth, and the viral code is incompatible with it. The differences leave lambda as helpless as a Windows computer virus inside a Mac. The same fate will befall other viruses that attack. The people who made this bacterium and its new code believe the feature will make it immune to all viruses. They call it rE.coli-57. And they have big plans for it.
rE.coli-57 is being built in a laboratory at Harvard Medical School by a team led by a young biologist named Nili Ostrov. For the past five years Ostrov has obsessed over every detail of the bacterium’s genetic reconstruction, putting in grueling hours under the fluorescent lights of the wet lab. It is the most elaborate gene-editing project in history and was the subject of a 2016 landmark paper in Science that identified 148,955 DNA changes necessary to make the cell virus-proof. Ostrov’s team had completed 63 percent of them, she and her colleagues reported, and the beast was doing fine.
Three years later the rebuilt cell is almost ready. Sometime soon the scene just envisioned will take place with not just one but hundreds of viruses in a petri dish. If rE.coli-57 survives, it may forever change the relation between viruses and their prey—including us.
Viruses are incredibly abundant, with 800 million of them covering every square meter of this planet. They vex us with illness, but they also torment industries that use cells to manufacture products from yogurt to pharmaceuticals. The biotech giant Genzyme (now part of Sanofi), which uses bacteria to make drug molecules, lost half its market value after a 2009 virus infection in its Allston, Mass., plant sabotaged its production line, triggering critical pharmaceutical shortages. Viruses are also an expensive scourge in the dairy industry, which employs bacteria to ferment cheese and yogurt—these products have to be dumped when the bacteria are hit by viral contamination. A virus-proof bacterium could be a billion-dollar bug.
Such a cell could also open up a new world of designer medicines. “If we want to make fancy antibodies and fancy protein drugs, we need to incorporate different chemistry into them,” Ostrov says. “That would be a game changer for drug companies.” All natural proteins are built from the same 20 amino acids, but rE.coli-57’s altered operating system would allow it to build new proteins using exotic amino acids, just as new LEGO pieces expand what can be built with the basic starter set. Designer proteins could target diseases such as AIDS or cancer with exquisite precision.
More controversially, rE.coli-57’s success could be a step toward making human cells virus-proof by rendering their DNA impervious to viral hijacking. That achievement would be invaluable to medical research, which suffers from viral infection of human cell lines in lab dishes that are used to develop and test therapeutic medicines. Skeptics, however, doubt recoded cells would function like “normal” ones, making them unreliable test beds. The idea also alarms those who fear such recoding puts us a little closer to creating human beings with designer DNA. (No one involved in the project has proposed designing people.) Just to recode one human lab-dish cell would be extraordinarily complicated because the human genome is 3.2 billion letters long, 800 times larger than E. coli’s. But rE.coli is an essential and mind-blowing first step.
Code breakers
Recoding defeats viral invaders because it alters the language a cell employs to make proteins, which are the molecules that all life uses to get anything done in the world. Proteins are made of smaller units known as amino acids, and each amino acid has a three-letter DNA code made of some combination of the four DNA bases: A, C, G and T. For instance, TGG means tryptophan, and CAA means glutamine. These three-letter codes are called codons, and every gene is simply a linear sequence of them.
The protein making happens when that sequence gets sent to cellular factories, ribosomes, where the codons pair up with molecules called transfer RNAs (tRNAs). Each tRNA has one end that binds to a particular codon and another that binds to one and only one kind of amino acid. As the sequence of codons moves through the protein assembly line, the tRNAs string together the amino acids until the protein is complete.
But there is an important peculiarity in this system: it has a lot of redundancy. There are 64 codons because there are 64 three-letter combinations of A, C, G and T. But there are only 20 amino acids. That means there are multiple codes for most of the amino acids. AGG stands for arginine, for example, but so does CGA. Some amino acids have six codons.
Back in 2004, George Church, a Harvard geneticist and Ostrov’s boss, began to wonder if all these codons were absolutely necessary. What if every AGG in the E. coli genome was changed to CGA? Because both code for arginine, the bacterium would still build all its normal proteins. But—and this is a key point—if the tRNA that pairs with AGG was also eliminated from the cell, then the AGG codon would be a dead end in the protein-building process.
As Church thought about the implications of getting rid of certain tRNAs, he had an epiphany. “I realized that this would make the cells resistant to all viruses,” he says, “which would be a potential very big bonus.” Viruses such as lambda reproduce by getting a cell to read viral genes and build proteins using those sequences. But if the tRNA for AGG is deleted from the cell, then every viral gene that includes an AGG codon will get stuck awaiting a tRNA that no longer exists, and no viral protein will be completed.
Viruses evolve furiously; Church suspected they would quickly work around a single vanished tRNA. But if enough codons and tRNAs were eliminated, it would be virtually impossible for a virus to spontaneously hit on the right combination of mutations to use the revised code. E. coli had seven codons that were relatively rare. They occurred in all 3,548 of its genes, an average of 17 times per gene. If all the corresponding tRNAs were eliminated, a virus would need to develop about 60,000 new sequences, each one calling for the right substitute codon in exactly the right spot. And that was just not going to happen.
In 2004 this scenario was just idle thought. It was hard enough to change a single gene in an organism; editing the thousands of genes necessary to eliminate every instance of certain codons was impossible. But by 2014 technological breakthroughs put doing so just on the edge of imaginable. So Church started looking for someone with the drive and organizational skills to tackle the largest gene-editing project in history.
That was when Ostrov arrived in his lab as a postdoctoral researcher. If Church was the architect of rE. coli-57, Ostrov became the engineer and general contractor. Ostrov had a lot of molecular construction experience. She grew up in Israel and attended Tel Aviv University, where she modified a protein by adding a few amino acids that bound a metal particle. When several of these modified proteins snapped together, they formed a nanowire that could carry current. “That was awesome,” Ostrov recalls. “I thought, wow, we can use biology to make useful things.” Later, at Columbia University, she earned her Ph.D. by engineering baker’s yeast to produce red pigment when it detected disease-causing microbes; the project earned a Grand Challenge Exploration award from the Bill & Melinda Gates Foundation for its use in detecting cholera.
It was an impressive résumé, but Church’s project was exponentially more difficult. The seven codons to be eliminated appeared 62,214 times in the E. coli genome. Recoding them all required making 148,955 changes to the DNA. There had been a lot of headlines about fast and easy gene editing, but no gene-editing tool was capable of making anywhere near that many changes.
Breakthroughs in DNA synthesis, however, pointed to another solution: build a recoded E.coli genome from scratch. DNA can be produced biochemically in special DNA printers, which work like an inkjet printer spraying As, Cs, Gs and Ts. Today’s DNA-synthesis companies can reliably make pieces of DNA up to about 4,000 letters long.
Credit: Campbell Medical Illustration
Around 2015 Ostrov’s team downloaded the standard E. coli genome, a long string of four million letters, from a database and put it on a computer. Then the researchers went through the entire sequence, changing all 62,214 instances of the seven rare codons to synonymous ones. (For safety, they also changed genes to make the bacterium dependent on a synthetic amino acid supplied in its nutrient broth. That synthetic molecule does not exist in nature, so the bacterium would die if it ever escaped the lab.) The result was the new rE.coli-57 genome scrolling across a computer screen. The scientists then divided its four million letters into 4,000-letter pieces with overlapping ends and sent the files to a DNA-synthesis shop. “We cut it on the computer,” Ostrov says, “literally like a Word document.” The company printed the DNA and sent it back by FedEx. The team assembled those 4,000-letter pieces into 87 large fragments of 50,000 letters each, which is about 40 genes.
Those fragments were just DNA, of course, and DNA is just code. A cell is needed to bring that code to life, and no one knows how to build one of those completely from scratch. Instead Ostrov took a piecework approach. She started with colonies of normal E. coli and slowly replaced each piece of their genome with a recoded fragment, one at a time, testing after every transplant to see if the patient survived.
Rebuilding a cell
On the long, black benches of the Church lab, amid centrifuges, vortex mixers, racks of pipettes and stacks of petri dishes, Ostrov’s team grew 87 colonies of normal E. coli in an incubator the size of a dormitory fridge, inserted a different 50,000-letter recoded fragment into groups of microbes, then waited to see if they would live. She did not get her hopes up. Perhaps evolution had chosen its codons for reasons that had escaped human understanding.
Surprisingly, most colonies did well. Only 20 of the revised segments stopped microbes from growing. But that was 20 too many. For rE.coli-57 to be virus-proof, all the recoded sections had to work. “First, we tried to narrow it down to which specific gene didn’t work,” Ostrov says. “We broke up the 40-gene segment into two 20-gene versions and tested those. Then we narrowed it down to four genes that might be the problem. Then one gene. And then we figured out which codon might be the problem.”
As it turned out, most of the trouble came from DNA printing errors. In other words, the sequences of DNA Ostrov’s team received were not exactly what it had ordered—a common issue in DNA synthesis until very recently. Ostrov went back to the company and got new error-free sequences. After the bad DNA was replaced, more than 99 percent of the redesigned genes worked. Recoding, it seemed, was not a crazy idea.
But there was a handful of remaining problems that seemed to be real issues with protein or DNA function, not quality control at the printer. Ostrov had to figure out what evolution knew that she did not. Why would changing to a synonymous codon, which coded for the exact same amino acid, kill or damage the organism?
Troubleshooting these spots was like blazing a trail through a wilderness for which there was no map. For example, the reproduction rate in bacteria with a recoded section 21 slowed to a crawl. Why? Because there was no scientific literature on these recoded DNA stretches to guide Ostrov—her team was the first to reshape them—she carefully analyzed the performance of all the genes in the section, comparing their products with those in normal bacteria. She found five linked genes that were intact but that, for some reason, were not doing anything.

Biologist Nili Ostrov and her colleagues at Harvard University have created rE.coli-57, an otherwise normal E. coli bacterium that has nearly 150,000 DNA changes throughout its genome intended to make it virus-proof. Credit: Steph Stevens
It turned out to be a problem with the genetic equivalent of an on/off switch. Genes are preceded by sequences of DNA called promoters that control whether the gene is active or not. In higher life-forms, promoters and genes are clearly delineated, with obvious starting and ending points, but sometimes bacterial genes overlap; the DNA sequence at the end of one gene actually doubles as the beginning of the next. Ostrov found that a DNA sequence in a gene called yceD was doing double duty as the promoter, the switch, for the five genes that followed. By recoding yceD, she had accidentally turned them off. She changed three codons on yceD so their DNA more closely matched the design of a known strong promoter. The output of the five genes surged, and the bacteria began reproducing normally.
Ostrov’s team had an even tougher challenge with recoded section 44, which had killed its colony entirely. The researchers narrowed the problem area down to a gene called accD that bacteria use to make fatty acids. The recoded cells were not making any accD protein at all. Ostrov ran a design analysis on the recoded gene and guessed that the problem was right at the beginning of its sequence. In DNA, As and Ts naturally bond, as do Gs and Cs. (In mRNA, the molecule that DNA uses to send code to the protein-making ribosome, a base abbreviated as U substitutes for the T, and it binds to the A with the same specificity.) If the letters are in a certain order—lots of As, say, followed by lots of Ts—the end of the molecule can fold on itself like sticky tape and gum up cellular machinery. On her computer, Ostrov redesigned the gene, revising 10 of its 15 recoded codons to other, synonymous ones that seemed less likely to form sticky folds. When she inserted the new piece of DNA into the bacteria, the colony sprang back to life.
So it has gone, one troubleshooting exercise at a time, the researchers tinkering with biology but thinking like mechanics, always following the design-build-test cycle of the engineer. Remarkably there have been no deal breakers. “So far we haven’t hit any impossible spots,” Ostrov says. “The code gives us a lot of wiggle room.”
Virus-proof
This year, after she added working genetic segments from one strain to working segments in another, Ostrov turned the original 87 strains into eight healthy lines, each with one eighth of the fully recoded genome. Every time the scientists combined segments, new incompatibilities arose and had to be troubleshot. But by early spring eight lines were quickly coming together into four, heading toward two. Sometime soon there will be one strain of 100 percent recoded rE.coli-57.
Once that strain is up and running, the final step will be to eliminate the tRNAs associated with the missing codons. The cell will be just fine because its genes will use synonymous tRNAs that still exist. But an incoming virus should not be fine at all. Its genes, which have not been reengineered, will have some codons that call for a tRNA that no longer exists. No tRNA means no amino acid at that point in the protein-building sequence, which stops assembly. No new viral protein, no new copies of the virus. The viral DNA remains marooned inside the cell, isolated, alone, unable to replicate and do any harm.
Ostrov plans to test this scenario in a microscopic version of the old film Mad Max Beyond Thunderdome, where a hero, trapped in an arena, has to beat a series of attackers. This arena will be a small glass container. The biologists will add lambda to a dish holding a healthy colony of rE.coli-57. Then they will step back and let the organisms battle to the death. If rE.coli survives, the researchers will add another bacteria-preying virus and, after that, another. It is difficult to envision a way for even the most gifted viruses to crack rE.colts elaborately altered code. But then again, no virus has ever been forced to try. Two organisms will enter—one will leave.
Ostrov is too cagey to commit to a date for the contest because she does not yet have the single completely recoded strain, but she believes she and her team are close. “Sooner rather than later,” she says. “Absolutely.” And she hints that a celebration with Brazilian cocktails that she likes may be coming shortly. “When it’s done, I won’t keep it quiet. I’ll call from the beach with one hand holding a caipirinha.”
Viral immunity alone will make rE.coli-57 worth celebrating, but the bacterium will also offer, as Ostrov and her colleagues put it in their Science paper, “a unique chassis with expanded synthetic functionality that will be broadly applicable for biotechnology.” In other words, the microbe will be a flexible platform for assembling new kinds of proteins.
That could be a boon for drug development. Many cancer and immunotherapy drugs are proteins that break down quickly in the body, but rebuilding them with exotic amino acids could greatly extend their life span. Church has already launched a start-up called GRO Biosciences (the acronym stands for “genomically recoded organism”) to design such therapeutics.
Altered life
A few years further out, the vision of recoded, virus-proof human cells looms. These cells could solve the ongoing problem of viral contamination of cultured human cell lines (such as the famed Henrietta Lacks cancer cells) used throughout medical research. In labs, lines of human cells are regularly employed as test beds to develop new medicines and ideas for therapies. But once viruses infect such cells, they are almost impossible to get rid of, so experiments get tossed out, and scientists have little choice but to start over. If the therapies could be developed faster, they would save lives. The Center of Excellence for Engineering Biology, a global collaborative effort with Church as a founding member, has named recoded human cells as its initial project. rE.coli-57 would clearly be a stepping-stone on that path.
Not surprisingly, the idea of redesigning the operating system of human cells alarms some critics. For one, the cells might not be reliable mimics of natural cells. And although the center’s scientists have never proposed doing anything with the cells beyond cultured cell lines, it might be possible to create a recoded human being who might also be virus-proof.
That would be bad, says Columbia University virologist Vincent Racaniello, who panned the idea on his scientific blog. “Multiple codons exist for a reason—among others they provide a buffer against lethal mutation,” he wrote. “Recoding the human genome in this way is not likely to be without serious side effects.”
None of the project scientists have suggested recklessly editing the DNA of a baby and seeing what happens, as occurred in China last year. What they do say is that a careful, transparent study of how recoded human cells behave could give us brand-new insights into the relation between us and many of our most injurious diseases. For all of our time on earth, we have been stuck with the 64-codon system—and the illness-causing viruses that take advantage of it. In a few years we may know if we have to accept that situation or not.
Ostrov is not a part of the center’s project—“Just to clarify, I do not recode human cells”—but says that it is important to explore the genetic unknown safely, in lab dishes. “Clearly, there’s a reason evolution has selected the codons it has. But we know there are other viable options,” she says. “By changing them, we get to investigate what happens. We’ll see what works and what doesn’t, and we’ll have a better understanding of the rules.” Knowing these principles may offer us a chance to improve some of the organisms that use them.

